The genome of eukaryotic cells in addition to information sequences (exons) contains non-coding inserts - introns. Therefore, before the start of protein synthesis, the molecules formed as a result of DNA transcription are spliced.
Definition of a concept
Splicing is the process of excising non-coding regions (introns) from transcriptional RNA followed by cross-linking of exons, which leads to the formation of a continuous sense sequence containing information about the primary structure of the protein. These manipulations are carried out by specialized nucleoprotein complexes - spliceosomes.
Splicing is one of the stages of the complex preparation of ribonucleic acid for translation, where the mRNA molecule serves as the matrix on the basis of which the amino acid chain is built in ribosomes according to the complementary principle.
RNA Processing
Processing refers to the totality of post-transcriptional modifications of RNA, which lead it to a species suitable for participation in protein synthesis. In other words, this is the process of converting a primary transcript (pre-mRNA) into messenger RNA. In addition to splicing, processing includes three more stages, which include capping, polyadeneling, and modification of the primary structure of RNA.
Capping is the formation at the 5´-end of RNA of a special nucleotide sequence called a cap (from the English cap - cap, cap). The process begins at the stage of transcription and is carried out at the expense of GTP energy. Cap protects mRNA from nucleases, and also acts as a signal peptide when translation is activated.
During polyadenelation, the poly (A) polymerase enzyme attaches the residues of adenylic acid to the 3 ´ end of the RNA, resulting in the formation of an oligo (A) fragment containing from 100 to 250 monomers (the so-called poly (A) tail). This design ensures the stability of mRNA in the cell.
Splicing is the third priority process, at the end of which RNA editing begins, including the modification of nucleotides (methylation, deamenation, etc.) and the insertion of additional nitrogen bases (most often uridyl bases) into the chain. At this stage, processing is completed, and mature mRNA leaves the cell nucleus in the cytoplasm.
Splicingosome
In the formation of splicingosomes, small nuclear RNAs (snRNAs) play a key role. Due to the high content of uridyl bases, they are also called uPHK (U1, U2, U3, etc.). Small ribonucleoprotein particles (snRNPs) are formed in combination with snRNA nuclear proteins, from which splicingosomes are formed - ellipsoid particles with a size of 25 × 5 microns and a sedimentation coefficient of 50-60S.
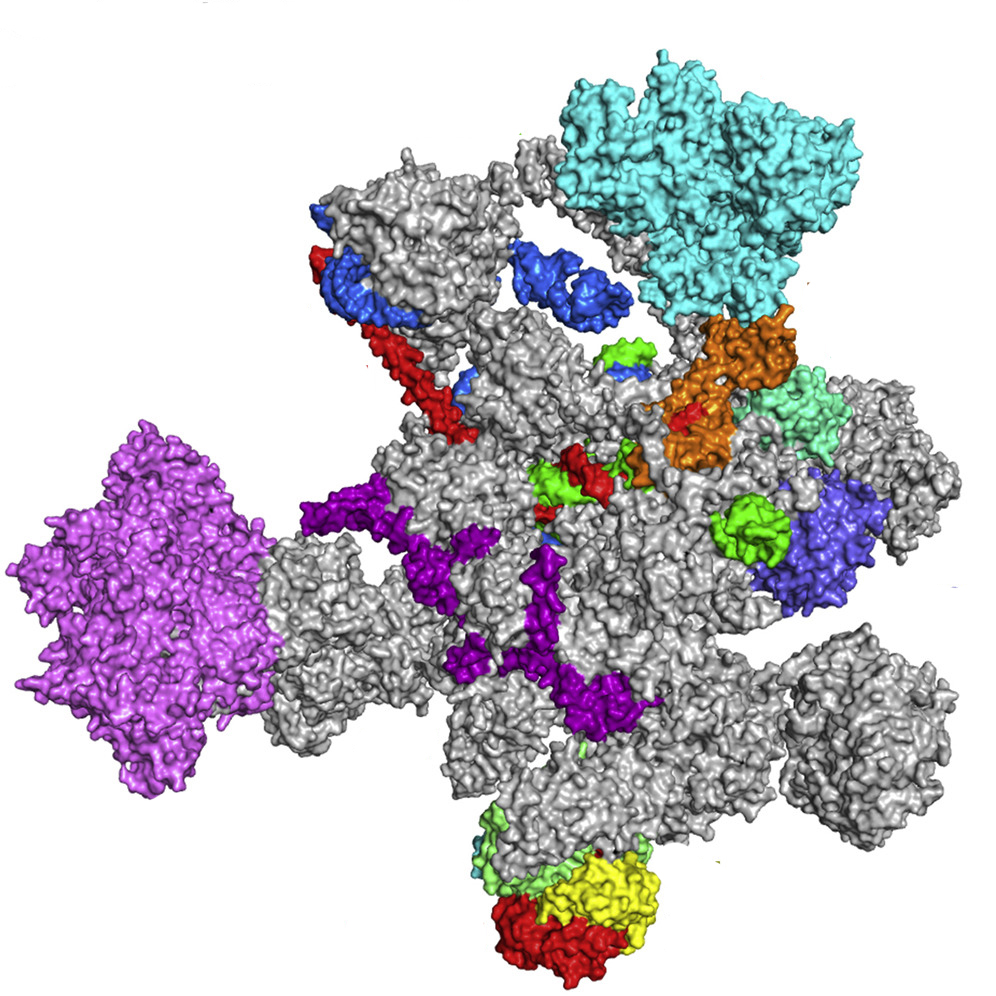
The mammalian spliceosome contains 6 varieties of snRNA (U1 – U6). These molecules are capable of complementary interaction with special conserved sequences at the ends of introns: GU and AG, called splicing sites. This leads to weaving and removal of the non-coding region from the RNA template sequence. The assembly of nucleoprotein particles into a single functional complex occurs directly during splicing. It is worth noting that the splicingosome itself does not cut and cross-link RNA regions; it only creates the conditions for a certain interaction between the chemical groups of nucleotides at the ends of exons and introns.
Intron removal mechanism
The ability of RNA splicing is largely determined by the intron structure feature: in addition to the consensus sequences (GU at the 5´ and AG at the 3´ ends), an adenyl nucleotide located in the highly degenerate purine-pyrimidine sequence (PyPyPuAPy ... ) The A nucleotide takes part in the formation of a lasso-type structure and is called the branch point. At the ends of exons are guanine nucleotides, which in the process of splicing form an incomplete GG bond.
The splicing mechanism is based on a change in the spatial configuration of the RNA molecule. Initially, various snRNAs complementarily bind to the intron GU and AG sites. At the same time, under the influence of structural proteins, nucleoprotein particles combine into a spliceosome, due to which the non-coding RNA region arches archfully, and the ends of exons come closer together to form an atypical hydrogen bond between guanine nucleotides.
As a result, the 3´-end of the first exon is adjacent to the adenyl nucleotide, and the phosphodiester bond at the boundary of the coding and non-coding sequences is destroyed, being replaced by a stronger complementary AG interaction. Thus, the intron forms a loop in the form of a lasso. Then, the hydroxyl group of the free end of the first exon attacks the 3´-site of splicing, cleaving the intron and binding to the second exon with the formation of whole RNA.
Alternative splicing
The coding regions of transcribed RNA can be crosslinked in various combinations to form alternative template sequences. In this case, it is possible to remove some exons or leave introns, which then participate in protein synthesis. The type of combination depends on the choice of splicing sites, which is regulated by various proteins. The mechanism of this process is currently poorly understood. Thus, alternative splicing is the process of forming different mRNAs based on one primary transcript.
Eukaryotes also have mechanisms for the formation of alternative pre-RNAs. These include the use of different promoters at the transcription stage and the change of polyadenelation sites.
The key role of alternative splicing is the ability to synthesize several protein isoforms on the basis of one semantic sequence, which increases the information capacity of DNA. So, in humans, due to this mechanism of 20 · 10 3 genes, about 10 5 types of proteins are encoded.